Welcome to the Mueller lab @ King's College London

Research Interests
Post-translational modifications of protein backbones
The polypeptide backbone makes up approximately 50% of every protein's mass. Originally thought to be inert, emerging evidence suggests that protein backbones are subject to a plethora of site-specific post-translational modifications. Similarly to well-studied modifications of amino acid side chains, backbone modifications can control protein structure and function. We are developing and applying a suite of chemical biology technologies to discover and manipulate protein backbone modifications. Our long-term goal is to elucidate when, where and how these fascinating modifications impact biological processes.
See also:
https://doi.org/10.1021/jacs.4c07103
Greg's collaboration with the Seebeck lab @ Uni Basel:
https://doi.org/10.1002/anie.202312104
Manuel's 2018 Biochemistry manifesto:
Molecular ageing
Proteins are subject to spontaneous modifications, contributing to senescence phenotypes across all kingdoms of life. For example, asparagine and aspartate residues can rearrange to isoaspartate, and long-lived proteins including eye lens crystalline contain significant amounts of isoaspartate. We aim to elucidate the biochemical, biophysical and cellular mechanisms of isoaspartate formation and explore the exciting possibility that this post-translational modification is harnessed as a molecular timer in biology.
See also:
Tianze and Luis' ChemRXiv preprint (with Fierz lab @ EPFL):
https://doi.org/10.26434/chemrxiv-2024-gh7fq
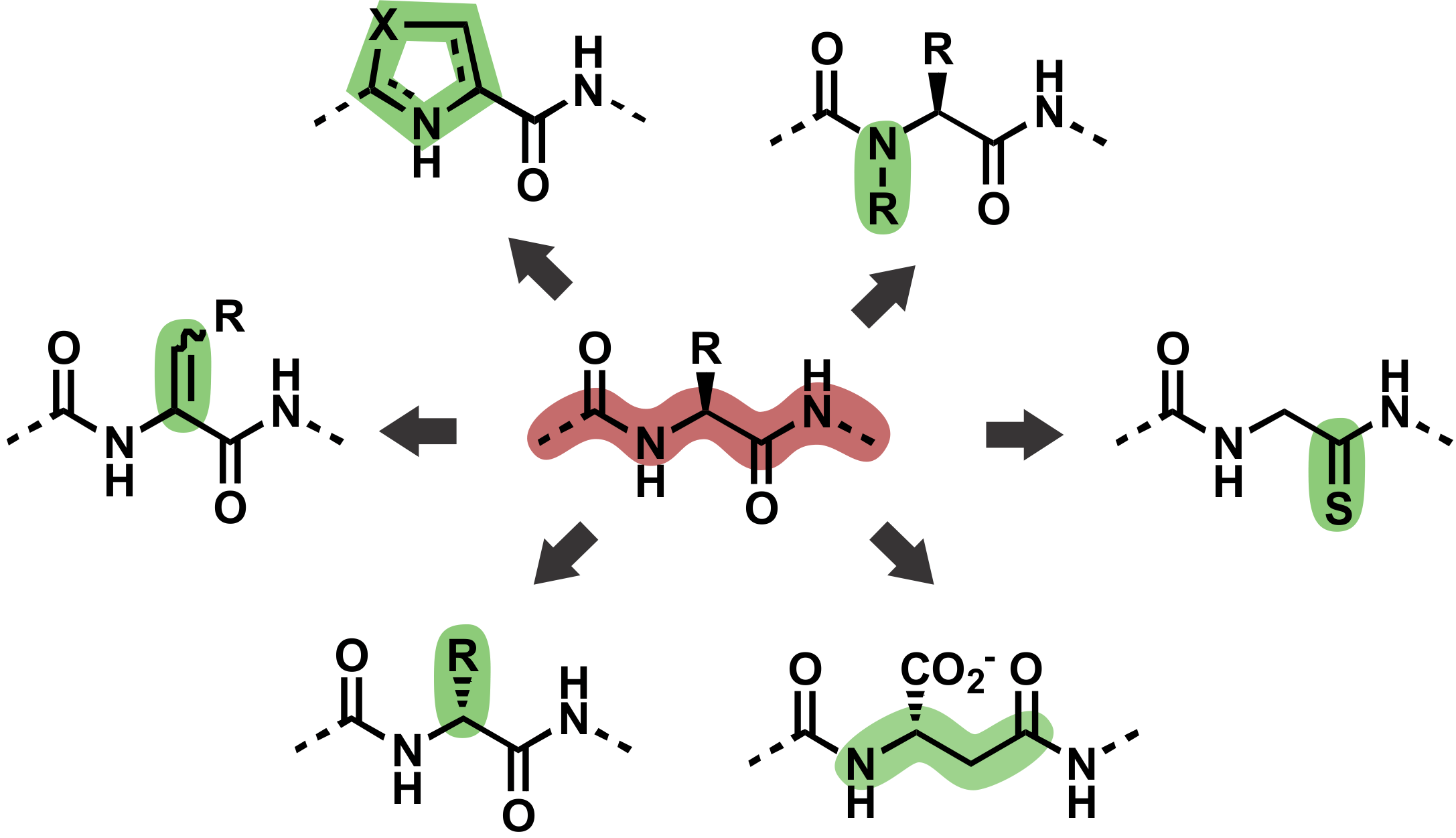
Post-translational modifications of the polypeptide backbone.
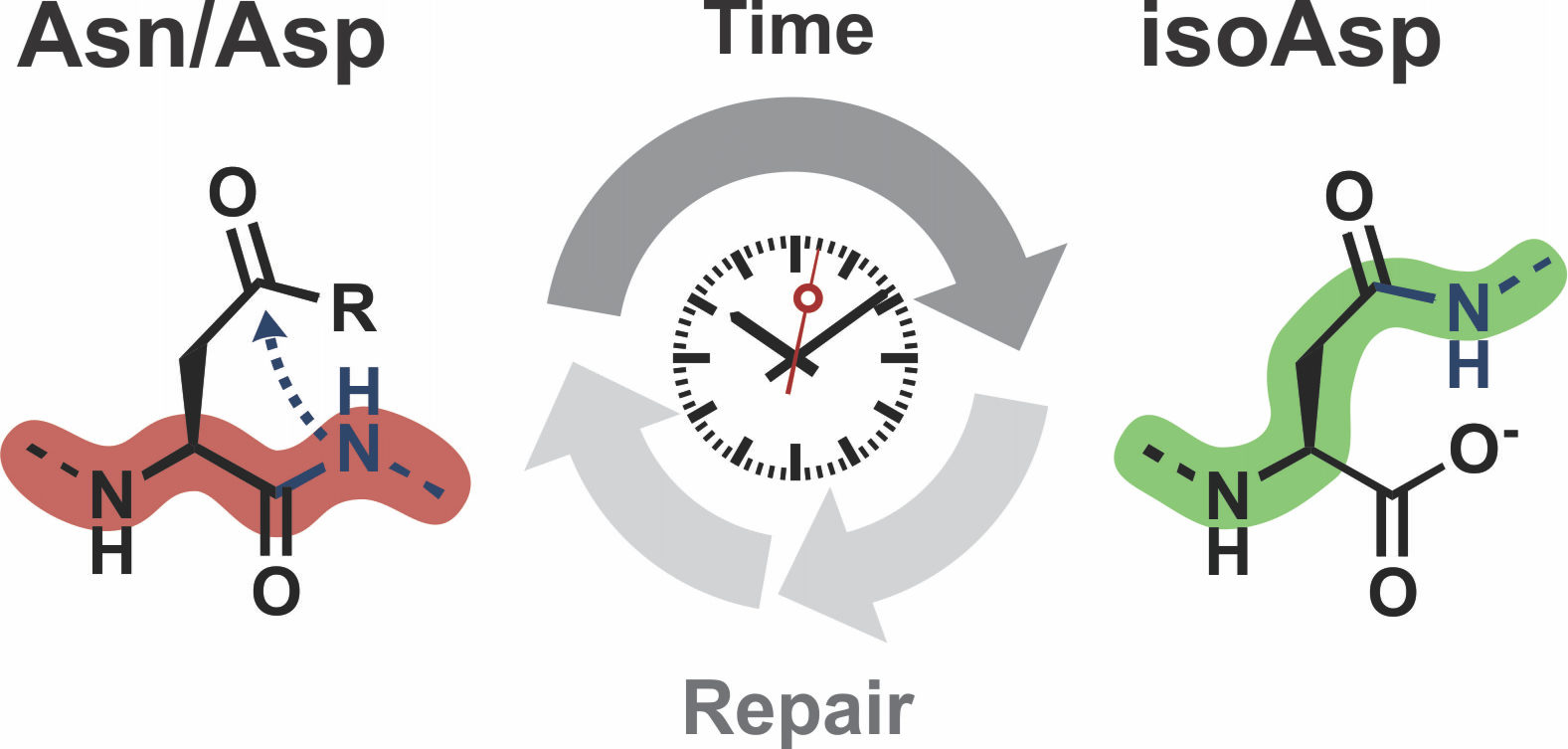
Iso-aspartate formation as a molecular timer.
Post-translational modifications in cellular life & death decisions
We employ cutting-edge protein semi-synthesis methods to prepare site-specifically modified proteins involved in cell death which enables us to investigate how these proteins are controlled by post-translational modifications. We are particular interested in understanding how the tumour suppressor protein p53 is activated in response to cell damage.
See also:
Sofia's & Karola's 2021 Chem Sci paper:
Jasmine's 2023 Chem Sci paper (with Tavassoli lab @ KCL):

Semi-synthesis of phospho-p53 via native chemical ligation.