Welcome to the Mueller lab @ King's College London

Visit Google Scholar for updated list of publications: Manuel M Mueller - Google Scholar
A Tailored Phospho-p53 Library Probes Antibody Specificity and Recognition Limitations
Hess M, Davis JH, Margiola S, Schneider S, Hicks T, Rai N, Müller MM
https://doi.org/10.1002/cbic.202500256

Semisynthesis of aged histone H4 reveals robustness and vulnerability of chromatin towards molecular "wear-and-tear"
Zhang T*, Guerra L*, Berlina Y, Wilson J, Fierz B, Müller MM
J. Am. Chem. Soc. in press (2025)
https://doi.org/10.1021/jacs.4c14136

Kinetic Resolution of Epimeric Proteins enables Stereoselective Chemical Mutagenesis
Ablat A, Lawton N, Alam R, Haybes BA, Hossain S, Hicks T, Evans SL, Jarvis JA, Nott TJ, Isaacson RL, Müller MM
J. Am. Chem. Soc. 146, 32, 22622–22628 (2024)
https://doi.org/10.1021/jacs.4c07103
Rapid Peptide Cyclization Inspired by the Modular Logic of Nonribosomal Peptide Synthetases
Ding Y, Lambden E, Peate J, Picken LJ, Rees TW, Perez-Ortiz G, Newgas SA, Spicer LAR, Hicks T, Hess J, Ulmschneider MB, Müller MM, and Barry SM
J Am Chem Soc 146, 16787-16801 (2024).
https://doi.org/10.1021/jacs.4c04711
FRET Sensor-Modified Synthetic Hydrogels for Real-Time Monitoring of Cell-Derived Matrix Metalloproteinase Activity using Fluorescence Lifetime Imaging
Yan Z, Kavanagh T, da Silva Harrabi R, Lust ST, Tang C, Beavil RL, Müller MM, Beavil AJ, Ameer-Beg S, da Silva RMP, Gentleman E
Adv Functl Mater 2309711 (2024).
https://doi.org/10.1002/adfm.202309711
Enzymatic Fluoromethylation as Tool for ATP‐independent Ligation
Peng J, Hughes GR, Müller MM*, Seebeck FP*
Angew Chem Int Ed. 63, e202312104 (2023).
https://doi.org/10.1002/anie.202312104
Angew Chem Int Ed. 62, e202305326 (2023)
https://doi.org/10.1002/anie.202305326
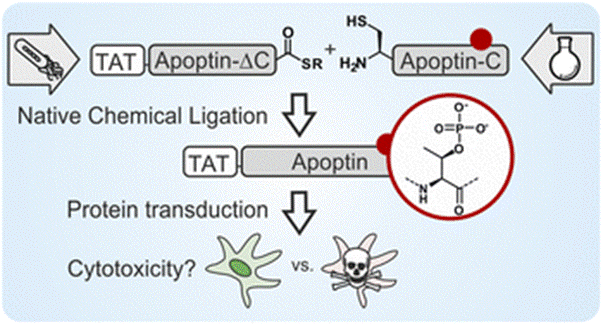
Semisynthesis reveals apoptin as a tumour-selective protein prodrug that causes cytoskeletal collapse
Wyatt J, Chan YK, Hess M, Tavassoli M*, Müller MM*
Chem Sci 14, 3881-3892 (2023).
Cysteine-Selective Modification of Peptides and Proteins via Desulfurative C−C Bond Formation
Griffiths RC, Smith FR, Li D, Wyatt J, Rogers DM, Long JE, Cusin LML, Tighe PJ, Layfield R, Hirst JD, Müller MM, Mitchell NJ
Chem Eur J 29, e202202503 (2023).
Semisynthetic ‘designer’ p53 sheds light on a phosphorylation-acetylation relay
Margiola S*, Gerecht K*, Müller MM
Chem Sci 12, 8563-8570 (2021).
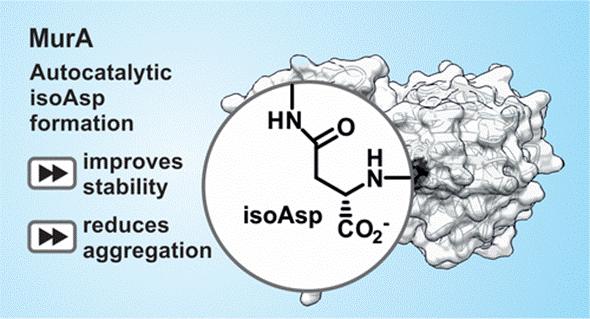
An unusually rapid protein backbone modification stabilizes the essential bacterial enzyme MurA
T Zhang, K Hansen, A Politis, MM Müller
Biochemistry 59, 3683–3695 (2020).
DOI: 10.1021/acs.biochem.0c00502
Cancer Treatment Goes Viral: Using Viral Proteins to Induce Tumour-Specific Cell Death
J Wyatt, MM Müller, M Tavassoli
Cancers 11, 1975 (2019).
Histone H3 tail binds a unique sensing pocket in EZH2 to activate the PRC2 methyltransferase
Jani KS, Jain SU, Ge EJ, Diehl KL, Lundgren SM, Müller MM, Lewis PW, Muir TW.
PNAS 116, 8295-8300 (2019).
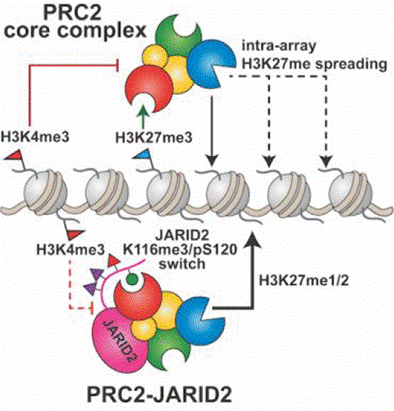
Nucleation and Propagation of Heterochromatin by the Histone Methyltransferase PRC2: Geometric Constraints and Impact of the Regulatory Subunit JARID2
Ge EJ, Jani KS, Diehl KL, Müller MM, and Muir TW
J Am Chem Soc 141, 15029-15039 (2019).
https://doi.org/10.1021/jacs.9b02321
HELLS and CDCA7 comprise a bipartite nucleosome remodeling complex defective in ICF syndrome
Jenness C, Giunta S, Müller MM, Kimura H, Muir TW, Funabiki H
PNAS 115 , E876-E885 (2018).
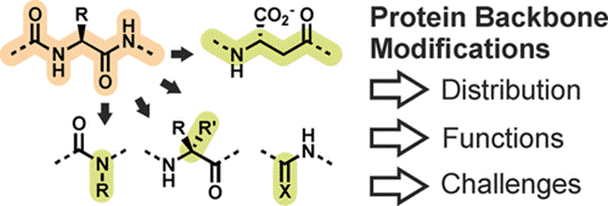
Post-Translational Modifications of Protein Backbones: Unique Functions, Mechanisms, and Challenges
Müller MM
Biochemistry 57, 177-185 (2018).
DOI: 10.1021/acs.biochem.7b00861
Chemical Protein Synthesis: Strategies and Biological Applications. In: Chemical and Biological Synthesis: Enabling Approaches for Understanding Biology (Westwood, Nelson eds), RSC press.
Müller MM (2018).
ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference
Dann GP, Liszczak GP, Bagert JD, Müller MM, Nguyen UTT, Wojcik F, Brown ZZ, Bos J, Panchenko T, Pihl R, Pollock SB, Diehl KL, Allis CD, Muir TW
A two-state activation mechanism controls the histone methyltransferase Suv39h1
Müller MM, Fierz B, Bittova L, Liszczak G, Muir TW
Nature Chem Biol 12, 188-193 (2016).
https://doi.org/10.1038/nchembio.2008
Evidence that ubiquitylated H2B corrals hDot1L on the nucleosomal surface to induce H3K79 methylation
Zhou L, Holt MT, Ohashi N, Zhao A, Müller MM, Wang B, Muir TW
Nat Commun 7, 10589 (2016).
doi:10.1038/ncomms10589
Histones: At the Crossroads of Peptide and Protein Chemistry
Müller MM and Muir TW
Chem Rev 115, 2296-2349 (2015).
doi:10.1021/cr5003529.
DNA-Guided establishment of nucleosome patterns within coding regions of a eukaryotic genome
Beh LY, Müller MM, Muir TW, Kaplan N, Landweber LF
Genome Res 25, 17271738 (2015).
doi:10.1101/gr.188516.114
Targeted histone peptides: insights into the spatial regulation of the methyltransferase PRC2 by using a surrogate of heterotypic chromatin
Brown ZZ, Müller MM, Kong HE, Lewis PW, Muir TW
Angew Chem Int Ed 54, 6457-6461 (2015).
doi:10.1002/ange.201500085
Strategy for “detoxification” of a cancer-derived histone mutant based on mapping its interaction with the methyltransferase PRC2
Brown ZZ*, Müller MM*, Jain SU, Allis CD, Lewis PW, Muir TW * = Co-first authors.
J Am Chem Soc 136, 13498–13501 (2014).
doi:10.1021/ja5060934
Building proficient enzymes with foldamer prostheses
Mayer C, Müller MM, Gellman SH, Hilvert D
Angew Chem Int Ed 53, 6978-6981 (2014).
doi:10.1002/anie.201400945
Accelerated chromatin biochemistry using DNA-barcoded nucleosome libraries
Nguyen UTT, Bittova L, Müller MM, Fierz B, David Y, Houck-Loomis B, Feng V, Dann GP, Muir TW
Nat methods 11, 834–840 (2014).
doi:10.1038/nmeth.3022
Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma
Lewis PW, Müller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, Allis CD
Science 340, 857-861 (2013).
doi:10.1126/science.1232245
Directed evolution of a model primordial enzyme provides insights into the development of the genetic code
Müller MM, Allison JR, Hongdilokkul N, Gaillon L, Kast P, van Gunsteren WF, Marlière P, Hilvert D
PLoS Genet 9, e1003187 (2013).
doi:10.1371/journal.pgen.1003187
Design, Selection, and Characterization of a Split Chorismate Mutase
Müller MM, Kries H, Csuhai E, Kast P, Hilvert D
Protein Sci 19, 1000-1010 (2010).
doi:10.1002/pro.377
A Rationally Designed Aldolase Foldamer
Müller MM, Windsor MA, Pomerantz MW, Gellman SH, Hilvert D
Angew Chem Int Ed 48, 922-925 (2009).
doi:10.1002/ange.200804996
Enhancing Activity and Controlling Stereoselectivity in a Designed PLP-Dependent Aldolase
Toscano MD, Müller MM, Hilvert D
Angew Chem Int Ed 46, 4468-4470 (2007).
doi:10.1002/anie.200700710
Patents
Muir TW, Nguyen UTT, Müller MM (2013). DNA Barcoding of Designer Mononucleosome and Chromatin Array Libraries for the Profiling of Chromatin Readers, Writers, Erasers, and Modulators thereof. Patent WO2013184930.